1. Standardizing
Data can contain all sorts of different values. It is hard to interpret when data take on any range of values. Therefore, we should convert the data into a standard format to make it easier to understand. The standard format of data refers to 0 means and unit variance. It is a simple process. For each data value, x, we subtract the overall mean of the data, μ, then divide by the overall standard deviation, σ.
The scikit-learn data preprocessing module is called sklearn.preprocessing. One of the functions in this module, scale, applies data standardization to a given axis of a NumPy array.
If for some reason we need to standardize the data across rows, rather than columns, we can set the axis keyword argument in the scale function to 1. This may be the case when analyzing data within observations, rather than within a feature.
2. Data Range
In this section, we will learn how to compress data values to a specified range.
Range Scaling
We can scale data by compressing data into a fixed range instead of standardizing it. One of the common use cases for this is compressing data into the range [0, 1].
It is a two-step process.
- For a given data value, we first compute the proportion of the value with respect to the min and max of the data
- We then use the proportion of the value to scale to the specified range.
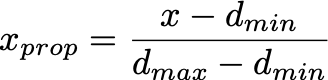
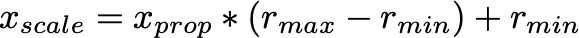
Range Compression in scikit-learn
In the previous chapter we used a single function, scale, to perform the data standardization, the remaining sections will focus on using other transformer modules.
The MinMaxScaler transformer performs the range compression using the previous formula. The default range is [0, 1].
The MinMaxScaler contains a function called fit_transform, which allows it to take in the input data array and then output the scaled data. The function is a combination of the object's fit and transform functions, where the former takes in an input data array and the latter transforms a (possibly different) array based on the data from the input to the fit function.
3. Robust Scaling
An outlier is a data point that is significantly further away from the other data points.
The data scaling methods from the previous two chapters are both affected by outliers. Data standardization uses each feature's mean and standard deviation, while ranged scaling uses the maximum and minimum feature values, meaning that they're both susceptible to being skewed by outlier values.
We can robustly scale the data, i.e. avoid being affected by outliers, by using the data's median and Interquartile Range (IQR). They are not affected by outliers. For the scaling method, we just subtract the median from each data value then scale to the IQR.
4. Data Imputation
Real-life datasets often contain missing values. If the dataset is missing too many values, we just don't use it. However, if only a few of the values are missing, we can perform data imputation to substitute the missing data with some other values.
-**There are several methods for data imputation. In scikit-learn, the SimpleImputer transformer performs four different data imputation methods.
- Using the mean
- Using the median
- Using the most frequent value
- Using a constant value
By using the strategy keyword argument when initializing a SimpleImputer object, we can specify a different imputation method.
Data imputation is not limited to those four methods.
There are also more advanced imputation methods such as k-Nearest Neighbors (filling in missing values based on similarity scores from the kNN algorithm) and MICE (applying multiple chained imputations, assuming the missing values are randomly distributed across observations).
5. PCA
When a dataset contains these types of correlated numeric features, we can perform principal component analysis (PCA) for dimensionality reduction (i.e. reducing the number of columns in the data array). It can extract the principal components of the dataset, which are uncorrelated set of latent variables that encompass most of the information from the original dataset.
we can apply PCA to a dataset in scikit-learn with a transformer, in this case the PCA module. When initializing the PCA module, we can use the n_components keyword to specify the number of principal components. The default setting is to extract m - 1 principal components, where m is the number of features in the dataset.


